GSoC ’25 Phase 2: Building OMOPCDMFeasibility.jl for Post-Cohort Feasibility Analysis
Introduction 👋
Hi everyone! I’m Kosuri Lakshmi Indu, a third-year undergraduate student majoring in Computer Science and a GSoC 2025 contributor. Over the past few months, I’ve had the opportunity to work with the JuliaHealth community, where I got a chance to learn, contribute, and get involved in projects focused on improving how we work with real-world health data.
As part of this project, I created and contributed to OMOPCDMFeasibility.jl, a Julia package within the JuliaHealth ecosystem. During Phase 2, I focused on designing and implementing practical tools for pre and post-cohort feasibility analysis on OMOP CDM data. This included developing utilities for cohort characterization, covariate profiling, and summary statistics—making it easier for researchers to explore, understand, and compare cohorts in real-world health datasets.
In this blog post, I’ll walk you through everything we accomplished in Phase 2, from the motivation behind these additions to examples of how they work in practice.
You can find my GSoC’25 Project Link
You can check out the project repository here: OMOPCDMFeasibility.jl
Official Documentation of OMOPCDMFeasibility.jl: Documentation
If you want, you can connect with me on LinkedIn and GitHub.
Background
What is OMOPCDMFeasibility.jl?
OMOPCDMFeasibility.jl is a Julia package designed to make post-cohort feasibility analysis on OMOP CDM data easy, reproducible, and robust. It provides a set of tools for characterizing cohorts, generating covariate profiles, and summarizing key statistics, all while supporting multiple SQL dialects and working seamlessly with the Julia data ecosystem.
The package is built to help researchers and data scientists quickly answer questions like:
How many patients meet my cohort criteria?
What are the demographic and clinical characteristics of my cohort?
How does my cohort compare to the overall database population?
Are my cohort definitions reproducible and robust across different database backends?
Project Description
Design and Structure
The core of OMOPCDMFeasibility.jl is organized in a clear, modular file structure, making it easy to navigate and extend:
OMOPCDMFeasibility.jl/
├── src/
│ ├── OMOPCDMFeasibility.jl # Main module file
│ ├── precohort.jl # Pre-cohort analysis: concept exploration, domain breakdown, eligibility checks
│ ├── postcohort.jl # Post-cohort analysis: cohort characterization, covariate profiling, summary stats
│ └── utils.jl # Utilities: person ID extraction, input validation, error handling, SQL supportprecohort.jl: Handles pre-cohort analysis, such as concept set exploration, domain breakdown, and eligibility checks. Use this module to analyze the distribution of concepts and get a sense of your data before defining a cohort.postcohort.jl: Focuses on post-cohort analysis, including cohort characterization, covariate profiling, and summary statistics. This is the main engine for generating demographic and clinical profiles.utils.jl: Contains utility functions for extracting person IDs, validating inputs, handling errors, and supporting dialect-agnostic SQL queries.
Deep Integration with the JuliaHealth Ecosystem
One of the greatest strengths of OMOPCDMFeasibility.jl is its deep and seamless integration with the JuliaHealth ecosystem and the OHDSI standards that underpin modern observational health research.
At the heart of this workflow is OMOPCohortCreator.jl. This foundational package not only enables the creation and management of cohorts from OHDSI JSON definitions, but also provides a rich set of covariate getter functions (such as GetPatientGender, GetPatientRace, GetPatientAgeGroup, and more). These functions are essential for stratified analyses and cohort profiling within OMOPCDMFeasibility.jl.
But the integration doesn’t stop there. OMOPCDMFeasibility.jl is designed to work hand-in-hand with a wide array of JuliaHealth tools, empowering users to build flexible, end-to-end pipelines for observational health research. This ecosystem approach ensures that your analyses are robust, reproducible, and scalable—making it.

Julia Health
Key Features and Examples
Pre-Cohort Analysis (precohort.jl)
The precohort.jl module is your toolkit for exploring and understanding your OMOP CDM data before you define a cohort. This is especially useful for feasibility checks and planning your study design. The main functions include:
analyze_concept_distribution: Examines how frequently specific OMOP concepts (such as conditions, drugs, or procedures) appear in your database. You can also stratify results by covariates (like gender or age group) to see how distributions differ across subpopulations.
using OMOPCDMFeasibility
using OMOPCDMCohortCreator: GetPatientGender, GetPatientAgeGroup
# Analyze the distribution of two concepts, stratified by gender and age group
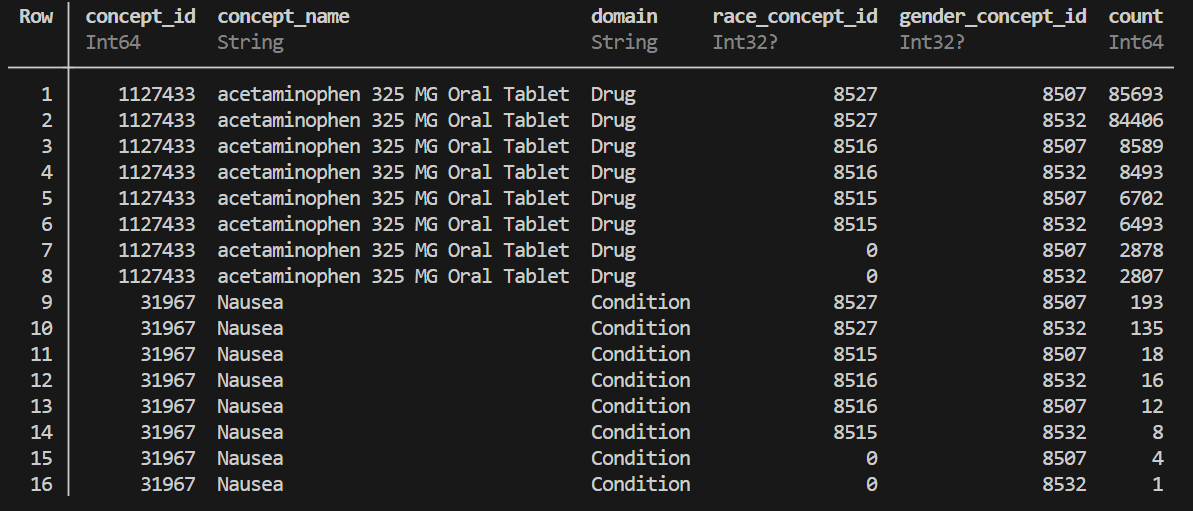
distribution = OMOPCDMFeasibility.analyze_concept_distribution(
conn;
concept_set=[
31967, # Condition: Nausea
1127433 # Drug: Acetaminophen
], covariate_funcs=[GetPatientGender, GetPatientRace],
schema="dbt_synthea_dev"
)
generate_summary: Produces summary statistics for a set of concepts, including the total number of patients, the number of eligible patients, and the coverage percentage. This helps you quickly assess the prevalence and relevance of your concepts of interest.
# Generate summary statistics for the same concepts
summary = OMOPCDMFeasibility.generate_summary(
conn;
concept_set=[
31967, # Condition: Nausea
1127433 # Drug: Acetaminophen
],
covariate_funcs=[GetPatientAgeGroup, GetPatientRace],
schema="dbt_synthea_dev"
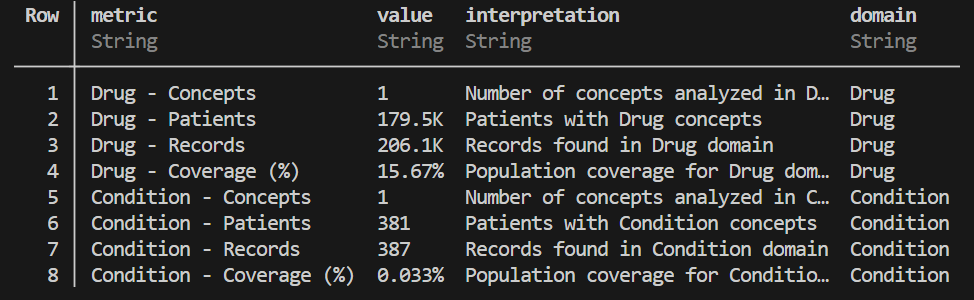
)generate_domain_breakdown: Breaks down your concept set by OMOP domain (e.g., Condition, Drug, Procedure), with options for raw or formatted output. This is useful for understanding the clinical context of your selected concepts.
# Break down the concepts by OMOP domain
domain_breakdown = OMOPCDMFeasibility.generate_domain_breakdown(
conn;
concept_set=[
31967, # Condition: Nausea
1127433 # Drug: Acetaminophen
],
schema="dbt_synthea_dev"
)
These functions help you understand the data landscape, check eligibility, and plan your cohort definitions before you commit to extracting a cohort.
Post-Cohort Analysis (postcohort.jl)
The postcohort.jl module is the core of post-cohort feasibility analysis. Here, you can deeply characterize your extracted cohort using flexible, composable functions.
Key Functions
create_individual_profiles: Generates demographic or clinical profiles for each subgroup in your cohort, based on any set of covariate functions (e.g., gender, race, age group). This helps you understand the makeup of your cohort at a granular level.create_cartesian_profiles: Produces multi-covariate (cartesian product) profiles, allowing you to analyze intersections of multiple covariates (e.g., “female patients aged (50-60) of a specific race”).
Input Options: cohort_df vs. cohort_definition_id
cohort_df: Pass a DataFrame of person IDs (and optionally, cohort start/end dates) if you have already extracted your cohort into Julia. This is ideal for in-memory analysis or when working with custom cohort tables.cohort_definition_id: Pass an integer ID to reference a cohort table stored in your database (created via OMOPCohortCreator.jl). This is useful for large-scale analyses or when working directly with database-resident cohorts.
Example usage:
using OMOPCDMFeasibility
using OMOPCDMCohortCreator: GetPatientGender, GetPatientRace, GetPatientAgeGroup
diabetes_concept_ids = [201826]
cohort_result = ConditionFilterPersonIDs(diabetes_concept_ids, conn)
cohort_ids = cohort_result.person_id
sample_cohort = DataFrame(
person_id = cohort_ids
)
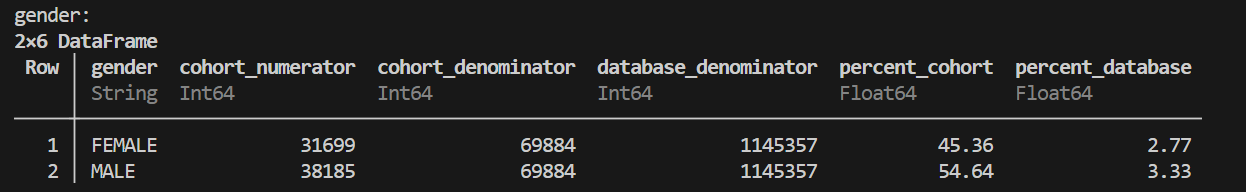
# Using a DataFrame of person IDs (cohort_df)
individual_demographics = OMOPCDMFeasibility.create_individual_profiles(
cohort_df=sample_cohort,
conn=conn,
covariate_funcs=[GetPatientGender, GetPatientRace]
)
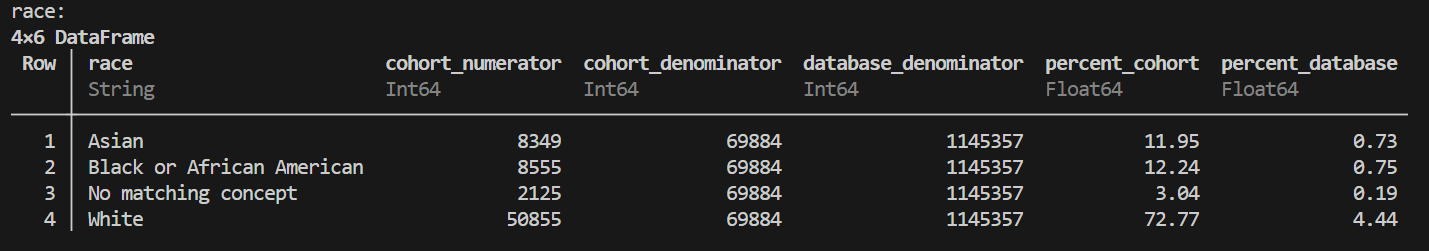
# Example 3: Multi-covariate (cartesian) profiling
cartesian_demographics = OMOPCDMFeasibility.create_cartesian_profiles(
cohort_df=sample_cohort,
conn=conn,
covariate_funcs=[GetPatientAgeGroup, GetPatientGender, GetPatientRace]
)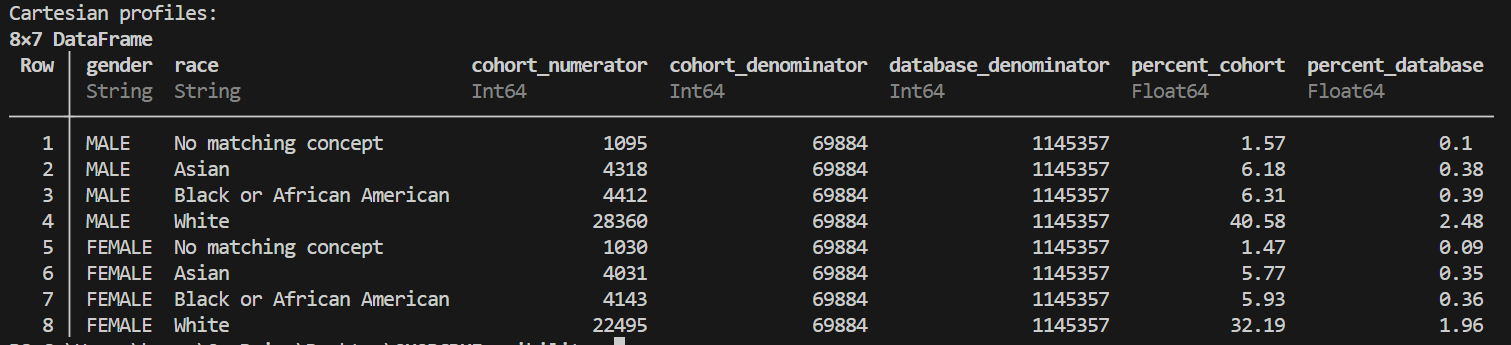
These functions allow you to flexibly analyze your cohort, whether you’re working with in-memory DataFrames or large database tables, and to stratify your results by any combination of covariates relevant to your database.
Future Scope
Looking ahead, here are some directions to further strengthen OMOPCDMFeasibility.jl and its integration within the JuliaHealth ecosystem:
Add more functions and mathematical/statistical analyses:
Extend the toolkit to support more complex cohort characterizations, outcome analyses, and advanced statistics.Use Genie.jl to create visual dashboards:
Build interactive dashboards for cohort exploration and reporting, making results more accessible to researchers and clinicians.Develop and utilize OMOP CDM datasets for testing:
Incorporate both real and synthetic OMOP CDM datasets to rigorously test the package’s features and ensure robust, generalizable performance across diverse data sources. This will help validate the toolkit and provide users with ready-to-use test cases.Ongoing maintenance and community-driven improvements:
Actively maintain the repository by addressing issues, updating dependencies, and refining documentation. Encourage community contributions to expand functionality, improve interoperability, and keep the package aligned with evolving
Final Thoughts
Thank you to everyone who made this project possible! Working on OMOPCDMFeasibility.jl has been an incredible learning experience. I’ve gained a deeper understanding of cohort analysis, reproducible research, and open-source development. I’m grateful for the support and mentorship I received, and I look forward to continuing to contribute to JuliaHealth and the open-source health data science community.
Acknowledgements
A big thank you to Jacob S. Zelko for being such a kind and thoughtful mentor throughout this project. His clear guidance, encouragement, and helpful feedback made a huge difference at every step. I’m also really thankful to the JuliaHealth community for creating such a welcoming and inspiring space to learn, build, and grow. It’s been a joy to be part of it.
Jacob S. Zelko: aka, TheCedarPrince
Note: This blog post was drafted with the assistance of LLM technologies to support grammar, clarity and structure.
Citation
@online{lakshmi_indu2025,
author = {Lakshmi Indu, Kosuri},
title = {GSoC ’25 {Phase} 2: {Building} {OMOPCDMFeasibility.jl} for
{Post-Cohort} {Feasibility} {Analysis}},
date = {2025-09-09},
langid = {en}
}